National Centre of Competence in Nanoscale Science, University of Basel
 |
Microfluidic networks project |
|
Jörg Ziegler (PhD stud), Martin Zimmermann (PhD stud), Dr. Marc Wolf (MD) More information: IBM RESEARCH LABORATORY, RUESCHLIKON/CH We show a proof-of-concept in which we combine our previously published concepts of micromosaic immunoassays (μMIAs) with self-regulating microfluidic networks (μFNs) to detect C-reactive protein (CRP) and other cardiac markers such as myoglobin (Mb) and cardiac Troponin I (cTnI). The μFNs are microfabricated in Si, have a well-defined surface chemistry, and are affixed to a bibulous material so as to self-regulate the displacement of an aliquot of liquid through the μFNs using capillary forces. An open section of the channels of the μFNs is covered with a hydrophobic poly(dimethylsiloxane) (PDMS) slab that acts as the substrate for a solid-phase immunoassay. Here, individual assays are conducted using independent channels. These assays are "sequential": series of samples, reagents, and buffers are displaced one after the other over the PDMS surface, and, as these assays are conducted under "microfluidic" conditions, they are fast to perform, very economical in their use of reagents, extremely integrated, and yield high-quality signals. The combinatorial character of μMIAs is exploited to optimize the assay parameters for detecting CRP. In particular, we found it optimal to deposit the capture antibody for CRP on PDMS at a concentration between 20 and 500 μg ml−1 in PBS in 1 min and to detect captured CRP in 2 min using a detection antibody having a concentration in PBS of 120 μg ml−1. With this method, CRP is quantitatively detected within 10 min in one microliter of human plasma down to concentrations of 30 ng ml−1, which suggests the possibility to detect CRP at clinically relevant concentrations for the management of coronary heart disease (CHD) and systemic inflammation. ( for details click here)
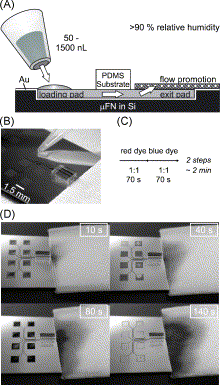
Method for exposing a PDMS substrate to solutions loaded sequentially into a μFN. (A) A solution initially placed in a loading pad spontaneously flows until it reaches a flow-promoting structure because of capillary forces. (B) This photograph shows a block of PDMS placed over the eight independent microchannels of this μFN. Each microchannel is linked to one loading pad, and the PDMS surface provides the substrate for the immunoassays. (C) Loading solutions consecutively into the same pad is denoted with arrows throughout this publication: Each arrow corresponds to an elementary step (filling, rinsing, blocking, etc.); compounds in the solution are indicated above the arrow, their concentration and the duration of the step below. Here, the two arrows refer to (D), in which all pads are loaded sequentially with a red and a blue solution and it takes
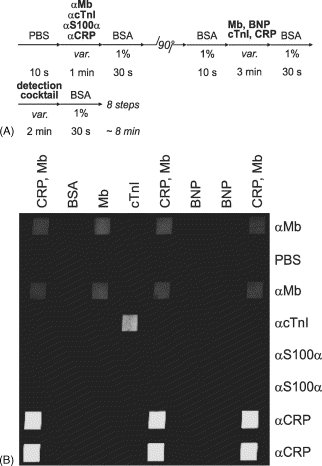
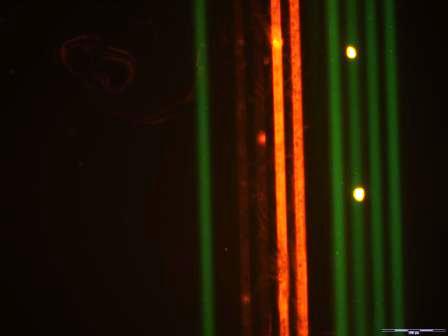
Simultaneous detection of cardiac markers in different samples using a μMIA. (A) Four capture Abs (αMb, αcTnI, αS100α, and αCRP) were deposited along horizontal lines of the μMIA and used to detect cardiac markers that were added to plasma samples of healthy subjects. Captured markers were detected by filling microchannels (vertical direction) with a cocktail of fluorescently-labelled Abs (αCRP-FITC, αBNP-Cy3, αS100α-Cy3, αMb-FITC and αcTnI-Cy3). (B) Fluorescence microscope image revealing the micromosaic of binding events. As expected, no signal for S100α and BNP was observed owing to the absence of either S100α in the plasma sample or αBNP capture Ab on the substrate.
|
||
|

 70 s for the solutions to move entirely from the loading pad to the flow-promoting structure (here a cleanroom tissue).
70 s for the solutions to move entirely from the loading pad to the flow-promoting structure (here a cleanroom tissue).